Data Types in R
Data types are the classification or categorization of data items. Data types represent a kind of value which determines what operations can be performed on that data. Numeric, character, and logical are the three most commonly used data types in R. Here's an overview of the different data types in R -
- Numeric
- Character
- Logical
- Factor
- Complex
- Raw
- Data and Time
Numeric data types are numbers stored in R objects. They can be integers such as 1, 2, 3, or floating-point numbers such as 1.2, 2.5, 3.7. They are used for mathematical calculations.
numeric_vector <- c(1.2, 2.5, 3.7)
Character data types are used to store strings of text. They are created by enclosing the text within double or single quotes.
character_vector <- c("apple", "banana", "cherry")
Logical data types are used to store logical values. They can be either TRUE or FALSE.
logical_vector <- c(T, F, T)
Factor data types are used to store categorical data. They can be ordered or unordered. They are created using the factor() function.
factor_vector <- factor(c("apple", "banana", "cherry"))
Complex data types are used to store complex numbers. They are created using the complex() function.
complex_vector <- complex(real = c(1, 2), imaginary = c(3, 4))
Raw data types are used to store raw bytes. They are created using the charToRaw() function.
raw_vector <- charToRaw(c("apple", "banana", "cherry"))
Data and time data types are used to store date and time values. They are created using the as.Date() and as.POSIXct() functions.
today <- Sys.Date() # Date
current_time <- Sys.time() # Date-time (POSIXct)
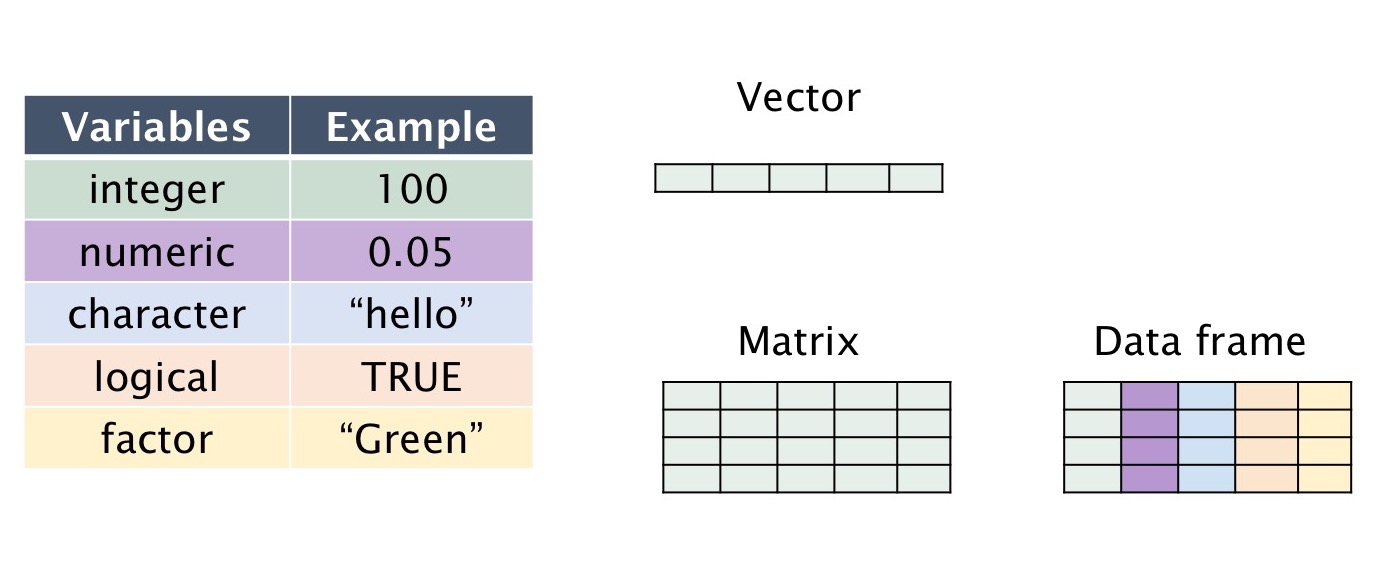
Data Structures in R
Data structures are fundamental components in computer science and programming. They are used to organize, store, and manipulate data efficiently. The choice of the right data structure depends on the specific problem you are trying to solve and the operations you need to perform on the data. Here's an overview of some common data structures used in R -
- Scalar
- Vector
- Matrices
- Data.frame
- List
- Tables
- Arrays
A scalar value refers to a single existing value, it could be in any data form (numeric, character, factor, logical, etc). For example, 1 is a scalar numeric value, 'a' is a scalar character value, F is a scalar logical value.
scalar_numeric <- 1
scalar_character <- "a"
scalar_logical <- F
A vector is the most basic data structure in R. It can hold elements of the same data type. Vectors can be numeric, character, logical, or other data types.
numeric_vector <- c(1.2, 2.5, 3.7)
character_vector <- c("apple", "banana", "cherry")
A matrix is a two-dimensional rectangular data set that contains elements of the same data type arranged in rows and columns. It can be created using a vector input to the matrix() function.
matrix(1:6, nrow = 2, ncol = 3)
A data frame is a two-dimensional data structure where each column can have a different data type. Data frames are commonly used for representing datasets. It can be created using the data.frame() function.
df <- data.frame(Name = c("Alice", "Bob", "Charlie"),
\t \t \t Age = c(25, 30, 22))
A list is a collection of objects that can be of different data types. It can be created using the list() function.
list(number_vector = 1:3,
\t character_vector = c("a", "b", "c"),
\t matrix_mine = matrix(1:6, nrow = 2, ncol = 3))
A table is a special type of data frame used for representing categorical data. It can be created using the table() function.
table(c("a", "b", "c", "a", "b", "c"))
An array is a multi-dimensional data structure that can hold elements of the same data type. It can be created using the array() function.
array(1:6, dim = c(2, 3))
Goals of Data Management in R
Data management is the process of ingesting, storing, organizing, and maintaining the data created and collected by an organization. It is a crucial part of the data science workflow. The goals of data management are -
- data quality
- Data Organization
- Data Documentation
- Data Cleaning
- Data Transformation
- Data Reproducibility
- Data Exploration
- Data Visualization
- Data Communication
Data quality refers to the accuracy, completeness, and consistency of data. It is important to ensure data quality because it affects the accuracy of the results of data analysis. Data quality can be improved by performing data cleaning operations such as removing duplicate values, handling missing values, and correcting inconsistent values.
Structure data in a clear and understandable manner. This includes organizing data frames, naming conventions, and creating data dictionaries.
Document data sources, data cleaning processes, and data transformations. Use comments and metadata to describe variables and datasets.
Data cleaning is the process of detecting and correcting corrupt or inaccurate records from a dataset. It is a crucial step in data management because it ensures data quality. Data cleaning can be performed using the dplyr package.
Data transformation is the process of converting data from one format or structure into another format or structure. It is a crucial step in data management because it ensures data quality. Data transformation can be performed using the dplyr package.
Data reproducibility is the process of reproducing the results of a data analysis. It is a crucial step in data management because it ensures data quality. Data reproducibility can be performed using the dplyr package.
Data exploration is the process of analyzing data to discover patterns, trends, and relationships. It is a crucial step in data management because it ensures data quality. Data exploration can be performed using the dplyr package.
Data visualization is the process of representing data in the form of charts, graphs, and maps. It is a crucial step in data management because it ensures data quality. Data visualization can be performed using the ggplot2 package.
Data communication is the process of presenting data in a clear and understandable manner. It is a crucial step in data management because it ensures data quality.
Packages of Interest
There are many packages available in R for data management. Here's an overview of some of the most commonly used packages -
- tidyverse
- Purr
- dplyr
- tidyr
- stringr
- lubridate
- readr
- readxl
- haven
- jsonlite
- xml2
- httr
- rvest
- pdftools
- magick
- ggplot2
tidyverse is a collection of packages for data management. It provides a set of functions for data manipulation, data visualization, and data communication. It is a part of the tidyverse collection of packages.
install.packages("tidyverse")
The packages available within tidyverse are as follows
dplyr & ggplot2 & tidyr & readr & purrr & tibble & stringr & forcats
Alternatively you can install many of the common packages manually, as below -
Purr is a package for list manipulation.
install.packages("purr")
dplyr is a package for data manipulation. It provides a set of functions for manipulating data frames. It is a part of the tidyverse collection of packages.
install.packages("dplyr")
tidyr is a package for data manipulation. It provides a set of functions for manipulating data frames. It is a part of the tidyverse collection of packages.
install.packages("tidyr")
stringr is a package for string manipulation. It provides a set of functions for manipulating strings. It is a part of the tidyverse collection of packages.
install.packages("stringr")
lubridate is a package for date manipulation. It provides a set of functions for manipulating dates. It is a part of the tidyverse collection of packages.
install.packages("lubridate")
readr is a package for data importation. It provides a set of functions for importing data. It is a part of the tidyverse collection of packages.
install.packages("readr")
readxl is a package for data importation. It provides a set of functions for importing data. It is a part of the tidyverse collection of packages.
install.packages("readxl")
haven is a package for data importation. It provides a set of functions for importing data. It is a part of the tidyverse collection of packages.
install.packages("haven")
jsonlite is a package for data importation. It provides a set of functions for importing data. It is a part of the tidyverse collection of packages.
install.packages("jsonlite")
xml2 is a package for data importation. It provides a set of functions for importing data. It is a part of the tidyverse collection of packages.
install.packages("xml2")
httr is a package for data importation. It provides a set of functions for importing data. It is a part of the tidyverse collection of packages.
install.packages("httr")
rvest is a package for data importation. It provides a set of functions for importing data. It is a part of the tidyverse collection of packages.
install.packages("rvest")
pdftools is a package for data importation. It provides a set of functions for importing data. It is a part of the tidyverse collection of packages.
install.packages("pdftools")
magick is a package for data importation. It provides a set of functions for importing data. It is a part of the tidyverse collection of packages.
install.packages("magick")
ggplot2 is a package for data visualization. It provides a set of functions for creating plots. It is a part of the tidyverse collection of packages.
install.packages("ggplot2")
Importing Data in R
Before we delve into data importation, it is important to understand the differences between absolute and relative paths.
- Absolute Path
- Relative Path
An absolute path is a path that points to the same location on one file system regardless of the working directory or combined paths. It is a complete path from start of actual filesystem from / directory.
read.csv("C:\Users\Documents\file.txt")
A relative path is a path that points to the same location on one file system relative to the current working directory. It is a path relative to the current working directory.
setwd("C:\Users\Documents") \br read.csv("file.txt")
In this case, we have set the working directory to "C:\Users\Documents" which allows us to specify the relative path without specifying the entire path.

Data can be imported into R from a variety of sources such as CSV files, Excel files, and databases. Here's an overview of some common data import methods in R -
- CSV Files
- dat Files
- Text Files
- SPSS Files
- Stata Files
- SAS Files
- Excel Files
- Database
- API
- Web Scraping
- Text Files
- JSON Files
- XML Files
- PDF Files
- Images
CSV files are text files that store tabular data in plain text format. They are commonly used for storing data in spreadsheets and databases. CSV files can be imported into R using the read.csv() function.
df <- read.csv("data.csv")
Since CSV files may often stored using different separtors. The following arguments may be of use -
df <- read.csv("data.csv", sep = ",", header = TRUE, stringsAsFactors = FALSE)
dat files are text files that store tabular data in plain text format. They are commonly used for storing data in spreadsheets and databases. dat files can be imported into R using the read.table() function.
df <- read.table("data.dat")
Text files are files that store text in plain text format. They are commonly used for storing data in spreadsheets and databases. Text files can be imported into R using the readLines() function.
df <- readLines("data.txt")
SPSS files are files that store data in SPSS format. They are commonly used for storing data in spreadsheets and databases. SPSS files can be imported into R using the haven package.
df <- haven::read_sav("data.sav")
Stata files are files that store data in Stata format. They are commonly used for storing data in spreadsheets and databases. Stata files can be imported into R using the haven package.
df <- haven::read_dta("data.dta")
SAS files are files that store data in SAS format. They are commonly used for storing data in spreadsheets and databases. SAS files can be imported into R using the haven package.
df <- haven::read_sas("data.sas")
Excel files are spreadsheet files that store tabular data in binary format. They are commonly used for storing data in spreadsheets and databases. Excel files can be imported into R using the read_excel() function.
df <- read_excel("data.xlsx")
A database is a collection of data stored in a computer system. It is commonly used for storing data in spreadsheets and databases. Databases can be imported into R using the dbConnect() function.
df <- dbConnect("data.db")
An API is a set of functions and procedures that allow the creation of applications that access the features or data of an operating system, application, or other service. It is commonly used for storing data in spreadsheets and databases. APIs can be imported into R using the httr package.
df <- httr::GET("https://api.data.com")
Web scraping is the process of extracting data from websites. It is commonly used for storing data in spreadsheets and databases. Web scraping can be performed using the rvest package.
df <- rvest::read_html("https://www.data.com")
Text files are files that store text in plain text format. They are commonly used for storing data in spreadsheets and databases. Text files can be imported into R using the readLines() function.
df <- readLines("data.txt")
JSON files are files that store data in JSON format. They are commonly used for storing data in spreadsheets and databases. JSON files can be imported into R using the jsonlite package.
df <- jsonlite::fromJSON("data.json")
XML files are files that store data in XML format. They are commonly used for storing data in spreadsheets and databases. XML files can be imported into R using the XML package.
df <- XML::xmlParse("data.xml")
PDF files are files that store data in PDF format. They are commonly used for storing data in spreadsheets and databases. PDF files can be imported into R using the pdftools package.
df <- pdftools::pdf_text("data.pdf")
Images are files that store data in image format. They are commonly used for storing data in spreadsheets and databases. Images can be imported into R using the magick package.
df <- magick::image_read("data.png")
Introducing the Piping Operator
The piping operator is a special operator that allows you to chain multiple operations together. It is a crucial part of the tidyverse collection of packages.

The piping operator, written as %>%, has been a longstanding feature of the magrittr package for R. It takes the output of one function and passes it into another function as an argument. This allows us to link a sequence of analysis steps. For example, we can use the piping operator to chain together multiple operations.
c(1,2,4,5,6,7,8) %>% sum() \br # Typical Code \br sum(1,2,3,4,5,6,7,8)
c(1,2,4,5,6,7,8) %>% sum() %>% mean() \br # Typical Code \br mean(sum(c(1,2,4,5,6,7,8)))
c(1,2,4,5,6,7,8) %>% sum() %>% mean() %>% round() \br # Typical Code \br round(mean(sum(c(1,2,4,5,6,7,8))))
To visualize this process, imagine a factory with different machines placed along a conveyor belt. Each machine is a function that performs a stage of our analysis, like filtering or transforming data. The pipe therefore works like a conveyor belt, transporting the output of one machine to another for further processing.
Dataframes vs Tibbles
| Data Frames | Tibbles | |
|---|---|---|
| Availability | Base R | Tidyverse |
| Column Name Conversion | Automatic | None |
| Data Type Coercion | Automatic | None |
| Row Names | Enabled by default | Disabled by default |
| Strict Printing | Less strict | Strict |
| Subsetting | Use of `[[]]` for columns and `[ ]` for rows | Consistent use of `[]` for both columns and rows |
| Data Type Columns | Not available | Includes a data type column |
Data frames and tibbles are both tabular data structures in R, but they have notable differences. Data frames, part of base R, automatically convert column names, can coerce data types, have row names by default, and offer more relaxed printing. Tibbles, associated with the tidyverse, do not alter column names, avoid data type coercion, exclude row names by default, and provide stricter and more informative printing. Moreover, tibbles allow consistent subsetting and include a data type column. The choice between data frames and tibbles depends on one's preference and the context of data analysis, with tibbles often favored for their user-friendliness within modern data analysis workflows.
# Creating a data frame
df <- data.frame(
\t name = c("Alice", "Bob", "Charlie"),
\t age = c(25, 30, 35),
\t gender = c("F", "M", "M")
)
print(df)
# Creating a tibble
library(tibble)
tb <- tibble(
\t name = c("Alice", "Bob", "Charlie"),
\t age = c(25, 30, 35),
\t gender = c("F", "M", "M")
)
print(tb)
Tibbles vs Tribbles
| Tibbles | Tribbles | |
|---|---|---|
| Availability | Part of the tidyverse | Introduced through `tribble` |
| Column Names | Preserved as is | Specified with `~` |
| Data Type Coercion | Not automatic | Not automatic |
| Printing | User-friendly, limited display | User-friendly, limited display |
# Creating a tibble
library(tibble)
tb <- tibble(
\t name = c("Alice", "Bob", "Charlie"),
\t age = c(25, 30, 35),
\t gender = c("F", "M", "M")
)
print(tb)
# Creating a tribble
library(tibble)
trb <- tribble(
\t ~name, ~age, ~gender,
\t "Alice", 25, "F",
\t "Bob", 30, "M",
\t "Charlie", 35, "M"
)
print(trb)
Dplyr Functions
| Category | Function | Utility |
|---|---|---|
| Data frame verbs (Rows) | arrange() | Order rows using column values |
| Data frame verbs (Rows) | distinct() | Keep distinct/unique rows |
| Data frame verbs (Rows) | filter() | Keep rows that match a condition |
| Data frame verbs (Rows) | slice() | Subset rows using their positions |
| Columns | mutate() | Create, modify, and delete columns |
| Columns | select() | Keep or drop columns using their names and types |
| Groups | group_by() | Group data by one or more variables |
| Data frames | bind_cols() | Bind multiple data frames by column |
| Data frames | bind_rows() | Bind multiple data frames by row |
| Vector functions | between() | Detect where values fall in a specified range |
| Vector functions | coalesce() | Find the first non-missing element |
In simple words, the dplyr package in R is like a set of powerful tools that help you easily and efficiently manipulate and transform data in data frames. It provides functions for filtering, sorting, summarizing, and modifying your data so that you can perform tasks like data cleaning and analysis with less code and more clarity. Think of it as a Swiss Army knife for working with data tables in R.
Arrange Function
The arrange() function is used to sort rows in ascending or descending order. It takes a data frame as input and returns a data frame with the rows sorted in ascending or descending order.
# Sort rows in ascending order
df <- arrange(df, age)
# Sort rows in descending order
df <- arrange(df, desc(age))
Distinct Function
The distinct() function is used to remove duplicate rows from a data frame. It takes a data frame as input and returns a data frame with the duplicate rows removed.
# Remove duplicate rows
df <- distinct(df)
Filter Function
The filter() function is used to select rows that match a condition. It takes a data frame as input and returns a data frame with the rows that match the condition.
# Select rows where age is greater than 30
df <- filter(df, age > 30)
Filtering with if_any(), if_all(), and between()
| Function | Description | Example |
|---|---|---|
| if_any() | Filter rows if any of the specified conditions are met. |
library(dplyr)
# Sample data frame
data <- data.frame(
Name = c("Alice", "Bob", "Charlie", "David"),
Age = c(25, 30, 22, 28),
Score = c(90, 75, 85, 95)
)
filtered_data <- data %>%
filter(if_any(c(Age, Score), ~ . >= 30))
|
| if_all() | Filter rows if all of the specified conditions are met. |
library(dplyr)
# Sample data frame
data <- data.frame(
Name = c("Alice", "Bob", "Charlie", "David"),
Age = c(25, 30, 22, 28),
Score = c(90, 75, 85, 95)
)
filtered_data <- data %>%
filter(if_all(c(Age, Score), ~ . >= 30))
|
| between() | Filter rows where a variable's value is within a specified range. |
library(dplyr)
# Sample data frame
data <- data.frame(
Name = c("Alice", "Bob", "Charlie", "David"),
Age = c(25, 30, 22, 28),
Score = c(90, 75, 85, 95)
)
filtered_data <- data %>%
filter(between(Age, 25, 30))
|
Mutate Function
The mutate() function is used to create new columns or modify existing columns. It takes a data frame as input and returns a data frame with the new or modified columns.
# Create a new column
df <- mutate(df, age_group = ifelse(age < 30, "young", "old"))
# Modify an existing column
df <- mutate(df, age = age + 1)
Using mutate() with across() and c_across()
| Function | Description | Example |
|---|---|---|
| across() | Apply a function or functions to multiple columns simultaneously within the mutate() function. |
library(dplyr)
# Sample data frame
data <- data.frame(
Name = c("Alice", "Bob", "Charlie", "David"),
Math_Score = c(90, 75, 85, 95),
English_Score = c(88, 92, 78, 86)
)
mutated_data <- data %>%
mutate(across(.cols = starts_with("Math"), .fns = ~ . * 1.1))
|
| c_across() | Combine values from multiple columns into a single column within the mutate() function. |
library(dplyr)
# Sample data frame
data <- data.frame(
Name = c("Alice", "Bob", "Charlie", "David"),
Math_Score = c(90, 75, 85, 95),
English_Score = c(88, 92, 78, 86)
)
mutated_data <- data %>%
mutate(Total_Score = c_across(starts_with("Score")))
|
Select Function
The select() function is used to select columns from a data frame. It takes a data frame as input and returns a data frame with the selected columns.
# Select columns by name
df <- select(df, name, age)
# Select columns by type
df <- select(df, where(is.numeric))
# Select columns by position
df <- select(df, 1:3)
Selecting with starts_with(), ends_with(), contains(), and matches()
| Function | Description | Syntax |
|---|---|---|
| starts_with() | Select columns that start with a specific prefix. | select(starts_with("prefix")) |
| ends_with() | Select columns that end with a specific suffix. | select(ends_with("suffix")) |
| contains() | Select columns that contain a specific substring. | select(contains("substring")) |
| matches() | Select columns based on regular expressions. | select(matches("regex_pattern")) |
| everything() | Select all columns. | select(everything()) |
| where() | Select columns based on a predicate function. | select(where(is.numeric)) |
Group By and Summarise Function
The group_by() function is used to group rows by one or more variables. It takes a data frame as input and returns a data frame with the rows grouped by the specified variables.
The summarise() function is used to summarise data by collapsing multiple values into a single value. It is often used in conjunction with the group_by() function to summarise data by groups.
library(dplyr)
# Sample data frame
data <- data.frame(
Name = c("Alice", "Bob", "Charlie", "David"),
Math_Score = c(90, 75, 85, 95),
English_Score = c(88, 92, 78, 86)
)
grouped_data <- data %>%
group_by(Name) %>%
summarise(Average_Math_Score = mean(Math_Score), Average_English_Score = mean(English_Score))
Vector Functions in dplyr
| Function | Description |
|---|---|
| between() | Detect where values fall in a specified range. |
| case_match() | A general vectorized switch() function. |
| case_when() | A general vectorized if-else function. |
| coalesce() | Find the first non-missing element among multiple vectors. |
| consecutive_id() | Generate a unique identifier for consecutive combinations of values. |
| cumall(), cumany(), cummean() | Cumulative versions of any(), all(), and mean() functions. |
| desc() | Sort data in descending order. |
| if_else() | Vectorized if-else function. |
| lag(), lead() | Compute lagged or leading values in a vector. |
| n_distinct() | Count unique combinations in a vector. |
| na_if() | Convert specified values to NA in a vector. |
| near() | Compare two numeric vectors to check if they are nearly equal. |
| nth(), first(), last() | Extract the nth, first, or last value from a vector. |
| ntile() | Bucket a numeric vector into n groups based on quantiles. |
| order_by() | A helper function for ordering in window functions. |
| percent_rank(), cume_dist() | Proportional ranking functions for a vector. |
| recode(), recode_factor() | Recode values in a vector. |
| row_number(), min_rank(), dense_rank() | Integer ranking functions for a vector. |
Using case_when() and case_match()
# Example for case_match()
library(dplyr)
data <- data.frame(Category = c("A", "B", "C", "D"))
mutated_data <- data %>%
mutate(Result = case_match(Category, "A" = "Alpha", "B" = "Beta", "C" = "Charlie"))
# Example for case_when()
library(dplyr)
data <- data.frame(Score = c(80, 95, 60, 75))
mutated_data <- data %>%
mutate(Grade = case_when(
Score >= 90 ~ "A",
Score >= 80 ~ "B",
Score >= 70 ~ "C",
TRUE ~ "D"
))
Using if_else() with mutate()
library(dplyr)
data <- data.frame(Score = c(80, 95, 60, 75))
mutated_data <- data %>%
mutate(Grade = if_else(Score >= 90, "A", "B"))
Using lag() and lead() with mutate()
library(dplyr)
data <- data.frame(Score = c(80, 95, 60, 75))
mutated_data <- data %>%
mutate(Lagged_Score = lag(Score), Leading_Score = lead(Score))
Joining Datasets
| Function | Description |
|---|---|
| anti_join() | Return rows from the left table that are not present in the right table. |
| full_join() | Return all rows from both tables. |
| inner_join() | Return rows that match in both tables. |
| left_join() | Return all rows from the left table. |
| right_join() | Return all rows from the right table. |
| semi_join() | Return rows from the left table that are present in the right table. |
| ID | Name |
|---|---|
| 1 | Alice |
| 2 | Bob |
| 3 | Charlie |
| 4 | David |
| ID | Score |
|---|---|
| 2 | 95 |
| 3 | 89 |
| 5 | 78 |
| 6 | 92 |

Using Join Functions
library(dplyr)
# Sample data frames
left_data <- data.frame(
Name = c("Alice", "Bob", "Charlie", "David"),
Age = c(25, 30, 22, 28)
)
right_data <- data.frame(
Name = c("Alice", "Bob", "Charlie", "David"),
Score = c(90, 75, 85, 95)
)
# Join data frames
joined_data <- left_data %>%
inner_join(right_data, by = "Name")
Tranform Datasets
The gather() function is used to transform a dataset from wide format to long format. It takes a data frame as input and returns a data frame with the columns gathered into key-value pairs.
library(tidyr)
# Sample data frame
data <- data.frame(
Name = c("Alice", "Bob", "Charlie", "David"),
Math_Score = c(90, 75, 85, 95),
English_Score = c(88, 92, 78, 86)
)
# Gather columns
gathered_data <- data %>%
gather(key = "Subject", value = "Score", Math_Score, English_Score)
Spread Datasets
The spread() function is used to transform a dataset from long format to wide format. It takes a data frame as input and returns a data frame with the key-value pairs spread across multiple columns.
library(tidyr)
# Sample data frame
data <- data.frame(
Name = c("Alice", "Bob", "Charlie", "David"),
Math_Score = c(90, 75, 85, 95),
English_Score = c(88, 92, 78, 86)
)
# Spread columns
spread_data <- data %>%
gather(key = "Subject", value = "Score", Math_Score, English_Score)
Separate Datasets
The separate() function is used to separate a column into multiple columns. It takes a data frame as input and returns a data frame with the column separated into multiple columns.
library(tidyr)
# Sample data frame
data <- data.frame(
Name = c("Alice", "Bob", "Charlie", "David"),
Age = c("25, 30", "30, 35", "22, 27", "28, 33")
)
# Separate column
separated_data <- data %>%
separate(Age, into = c("Age_1", "Age_2"), sep = ", ")
Unite Datasets
The unite() function is used to unite multiple columns into a single column. It takes a data frame as input and returns a data frame with the columns united into a single column.
library(tidyr)
# Sample data frame
data <- data.frame(
Name = c("Alice", "Bob", "Charlie", "David"),
Age_1 = c(25, 30, 22, 28),
Age_2 = c(30, 35, 27, 33)
)
# Unite columns
united_data <- data %>%
unite(Age, Age_1, Age_2, sep = ", ")
| Spread Data (Wide Form) | |||
| Name | Exam 1 | Exam 2 | Exam 3 |
|---|---|---|---|
| Alice | 85 | 88 | 78 |
| Bob | 90 | 92 | 85 |
| Charlie | 78 | 86 | 92 |
| Gathered Data (Long Form) | ||
| Name | Exam | Score |
|---|---|---|
| Alice | Exam_1 | 85 |
| Bob | Exam_1 | 90 |
| Charlie | Exam_1 | 78 |
| Alice | Exam_2 | 88 |
| Bob | Exam_2 | 92 |
| Charlie | Exam_2 | 86 |
| Alice | Exam_3 | 78 |
| Bob | Exam_3 | 85 |
| Charlie | Exam_3 | 92 |
Brief Introduction to R Markdown
R Markdown is a file format for making dynamic documents with R. An R Markdown document is written in markdown (an easy-to-write plain text format) and contains chunks of embedded R code. R Markdown files are designed to be used with the rmarkdown package. R Markdown files have the file extension “.Rmd”. When you render an R Markdown document, R Markdown converts your document into the desired output format.
Setting up a R Markdown file -
When you open a new R Markdown file in RStudio, a pop-up window appears that prompts you to select output format to use for the document. You can select from a variety of output formats including HTML, PDF, MS Word, and more. You can also select the default output format for all new R Markdown files in the R Markdown preferences.

R Markdown Syntax
R Markdown files are written using markdown, a lightweight markup language that is easy to read and write. Markdown is a way to style text on the web. You control the display of the document; formatting words as bold or italic, adding images, and creating lists are just a few of the things we can do with Markdown. Mostly, Markdown is just regular text with a few non-alphabetic characters thrown in, like # or *.
| R Markdown Syntax | Description |
|---|---|
# Header 1 |
Create a top-level heading (Header 1). |
## Header 2 |
Create a second-level heading (Header 2). |
*Italic* |
Italicize text using asterisks. |
**Bold** |
Make text bold using double asterisks. |
[Link](https://example.com) |
Create a hyperlink with the specified text and URL. |
 |
Embed an image in the document with alt text and image source. |
> Blockquote |
Create a blockquote for cited content. |
* List item 1 |
Add an unordered list item with an asterisk. |
1. Numbered item |
Include a numbered list item. |
--- |
Insert a horizontal rule (horizontal line). |
Inline |
Mark inline code using backticks. |
```Code block```
|
Create a code block for displaying and formatting code. |
Embedding R Code in R Markdown
R Markdown files can contain chunks of embedded R code. You can embed an R code chunk in an R Markdown file by using the chunk option in the RStudio toolbar. You can also use the keyboard shortcut Ctrl + Alt + I (Windows/Linux) or Cmd + Option + I (Mac).
| Option | Value | Description |
|---|---|---|
| eval | TRUE | Whether to evaluate the code and include its results |
| echo | TRUE | Whether to display code along with its results |
| warning | TRUE | Whether to display warnings |
| error | FALSE | Whether to display errors |
| message | TRUE | Whether to display messages |
| tidy | FALSE | Whether to reformat code in a tidy way when displaying it |
| results | "markup" | Output format for code results (e.g., "markup", "asis", "hold", or "hide") |
| cache | FALSE | Whether to cache results for future renders |
| comment | "##" | Comment character to preface results with |
| fig.width | 7 | Width in inches for plots created in the chunk |
| fig.height | 7 | Height in inches for plots created in the chunk |
About Me
I am a 2nd year PhD student in the Quanitative Methods Department at York University. Although my research primary revolves around adapting machine learning methodologies to Psychology, I take immense pleasure in improving the statistical literacy for everyone involved.
My research interests include = Machine Learning, Psychometrics, statistical pedagogy, and Data Science.
My hobbies include = Jiu Jitsu, Coding, Working Out, and Reading.
“Success is not final, failure is not fatal: It is the courage to continue that counts.” - Winston S. Churchill
